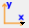
Data browsing frame#
The data browsing frame allows the visualization of one-dimensional and two-dimensional data. The frame is divided in two main parts: On the left, a directory explorer allows to browse the full file system and select files. On the right side, a data display allows to visualize data.
Files are selected by double-clicking on the respective file in the directory. For raw images, an additional selection widget to specify the data settings will be shown. Similarly, for hdf5 files, an additional selection widget to select the dataset in the file will be shown.
In the screenshot above, the data display is empty as no data has been selected. An example will be shown in the following sections.
The width of the directory explorer widget can be adjusted by using the dark grey handle between the two main widgets, to reduce or to enlarge the directory explorer, respectively. The draggable handle is highlighted with the green frame in the screenshot above.
Warning
The two main widgets have a defined minimum size. If the user drags the splitter further, the respective widget will be hidden. It can be enlarged again by capturing and dragging the dark slider from the edge towards the center.
Controls#
The controls are located on the left, above the directory explorer. Selecting or deselecting an option will directly update the directory explorer.
The options and control widgets are:
Show network drives
This option allows to show or hide linked network drives in the explorer view.
Sorting is case sensitive
This option toggles case sensitive sorting. If enabled, lowercase and uppercase names will be sorted and displayed separately.
Current directory
This widget both shows the current directory and allows to change the current directory by entering a new path. If the new path does not exist, the entry will be ignored. It is also possible to drag and drop directories or files from the file system into the entry field. This will automatically set the current directory to the directory of the dragged item. In the case of files, the file will also be opened, if it is a supported data format.
Filename filter
This field allows to filter the displayed files in the directory explorer. The asterix * can be used as a wildcard. For example, entering *.h5 will display only files with the .h5 extension. The filter is applied to files only and will not affect the display of directories, i.e. all directories will always be shown, regardless of the filter. The filter can be reset by deleting the entry in the field.
Reset filter
This button also resets the filename filter. It is equivalent to deleting the entry in the filter field.
Collapse all
This button collapses all directories in the directory explorer. It is equivalent to double-clicking on all top level entries.
Directory explorer#
The directory explorer is used to select the data to be displayed. The exact look and feel will depend on the used operating system and might be different from the screenshots shown here.
A single click on an item will just highlight the item but will otherwise be ignored. Double-clicking on a folder (or the arrow next to a folder) will expand or collapse the folder, depending on the folder’s current state. Double-clicking on a file will instruct pydidas to open the selected file. If the data format is readable and the file contains two-dimensional data, the content will be displayed in the ImageView widget. In case of hdf5 files, an additional selection field will be shown to select the data frame.
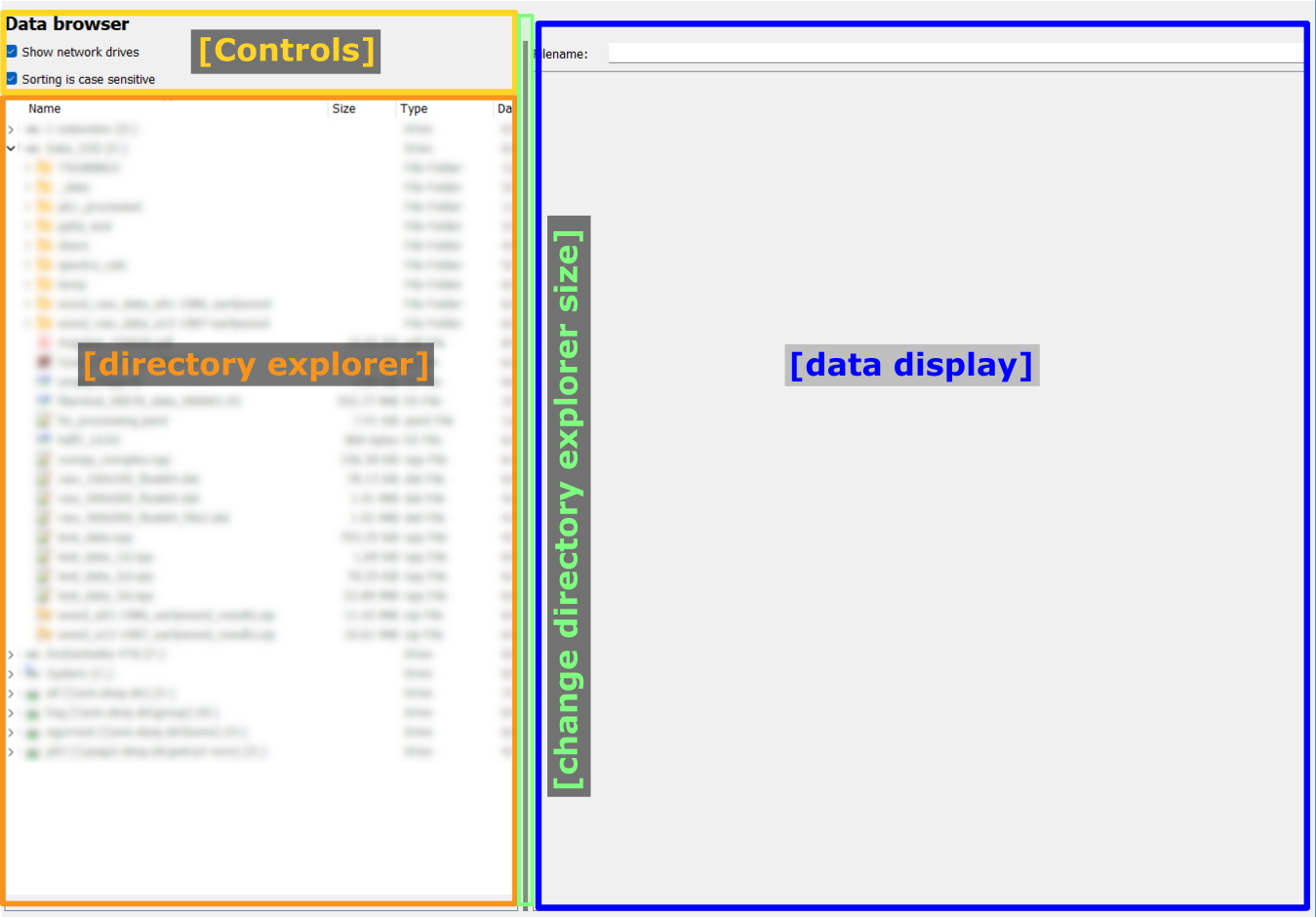
Data display#
The data display widget is shown on the right with the default 2D image view for multi-dimensional data.
Key elements are:
The filename display at the top: This widget shows the full path of the opened file.
Selection widgets for raw images and hdf5 files: These widgets allow to select the data settings for raw images and the dataset for hdf5 files.
The visualization widget: The selected data is displayed here.
The slice selection widget: Select the slicing in multi-dimensional datasets to select a two-dimensional slice for visualization.
The display modality selection: Select how to display the data.
Details for all elements are given below.
Note
The data display cannot process any metadata like axis labels or ranges. Only the raw data is displayed and only indices can be used to select.
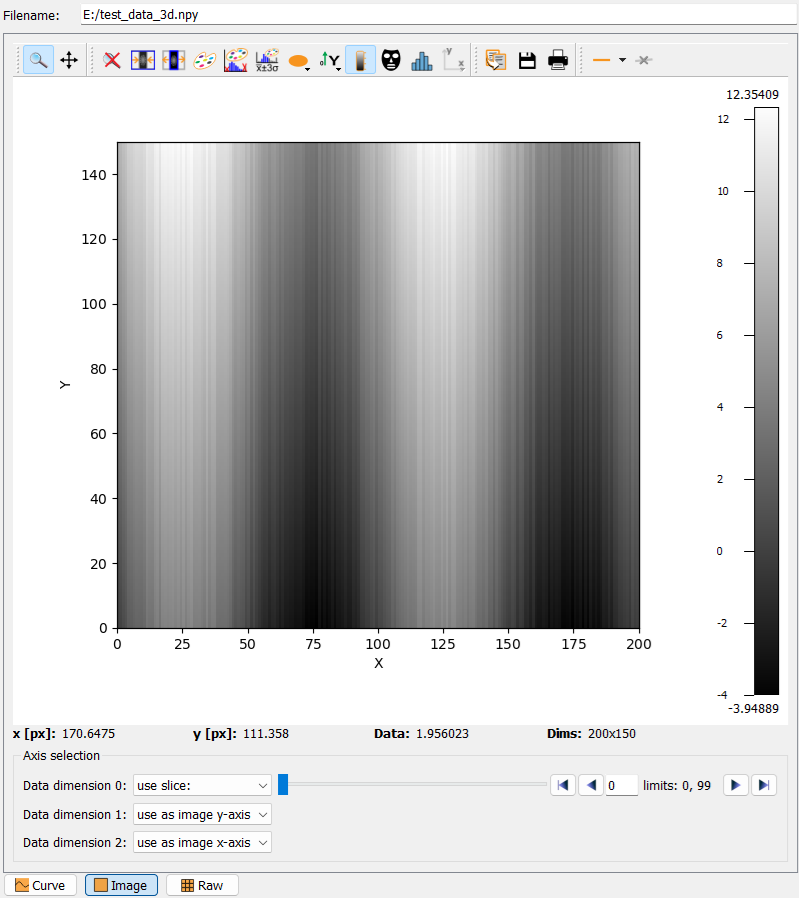
The hdf5 data selection widget#
The hdf5 data selection widget is shown below.

In the minimized view, it allows to open a window to display the hdf5 file structure (Hdf5 Browser Window), and a combo box to select the dataset to display. An additional button allows to show more dataset filter options. The full widget with all filter options is shown below:
The first row allows the user to select dataset filters for specific names. For example, the Eiger detector master file has a number of datasets for detector specific settings like offsets and calibrations for the different modules. If the respective box is ticked, these datasets will not be shown in the drop-down list. Additional filters for datasets can be set on their and minimum data dimension. Any changes to the filters will update the list of filtered datasets immediately.
To select a dataset, simply select the corresponding hdf5 dataset key from the drop-down list of the combo box. This will update the selection of the data frame.
Browsing multi-dimensional datasets uses the slice selection widget, which will be explained below.
The raw data selection widget#
Importing raw data files requires an additional selection of data type, image shape and header length (the header length is given in bytes). Settings all these values allows to correctly decode raw images. The respective widget is shown below:

Trying to decode raw data with wrong settings raises a warning message if the data cannot be imported.
A checkbox with Automatically load files with these settings allows to automatically apply these settings to a series of files with the same settings.
The Hide detailed options button allows to minimize the widget to a minimal size to increase the available space for the data display.
Display modality and slice selection#
At the bottom of the data display widget, the display modality and slice selection widgets allow to define how the data is displayed and which slice is shown.
An exemplary view is shown below:
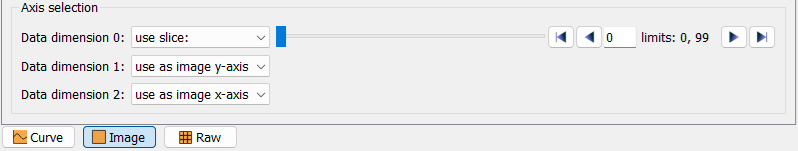
Depending on the data dimensionality, the modality selection will show different options:
Curve: Display a one-dimensional slice of the data as a line plot.
Image: Display a two-dimensional slice of the data as an image.
Raw: Display the raw data as a table.
The axis selection is required to specify which information to plot. For a curve view, one data dimension can be specified as curve y. The other axes will show a use slice option and a slider to select the slice to be plotted. For an image view, two axes can be selected to use as image y-axis and use as image x-axis, respectively. The other axes will show a use slice option and a slider. Similarly, for the raw view, two axes can be selected to use as row and use as column, respectively.
2D image view#
The PydidasPlot2d is a
subclassed silx Plot2d
with additional features useful in pydidas.
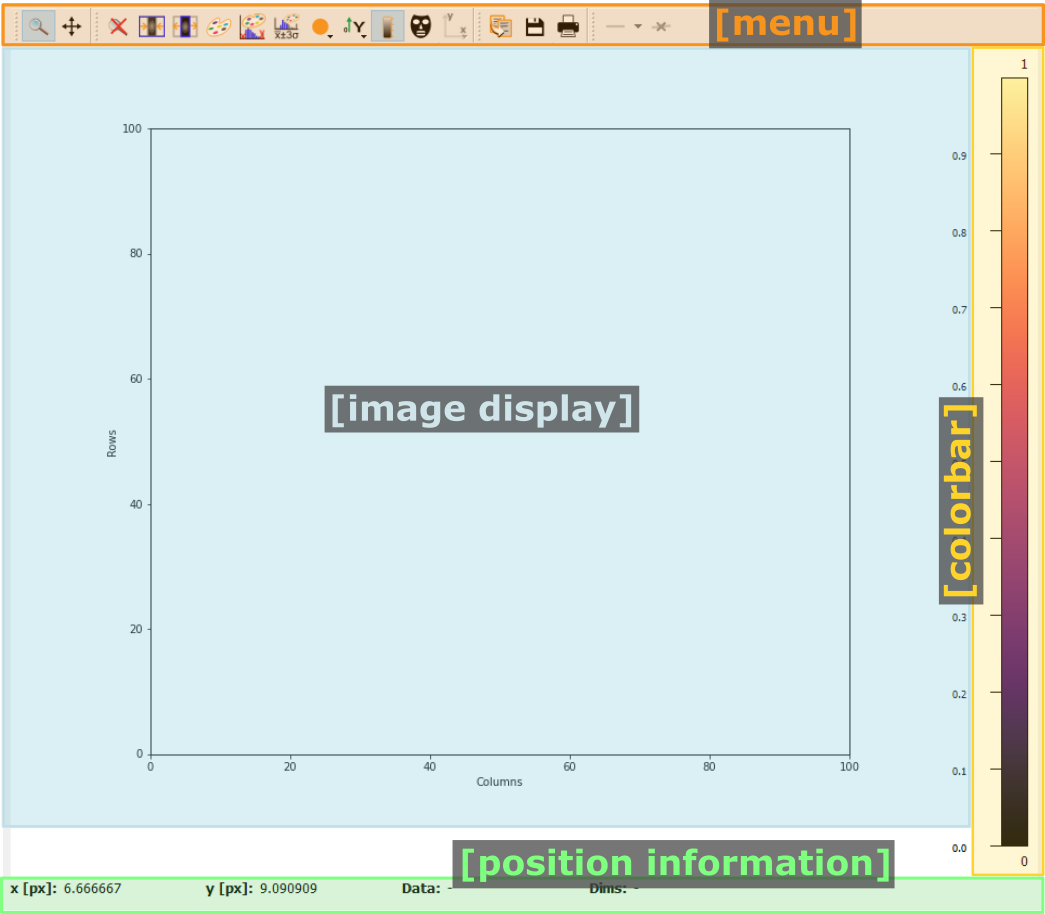
- The menu
The menu bar allows access to all generic silx and additional pydidas functionality. The detailed menu icons and actions are described below in the menu entries description.
- The image display
This widget shows the image data. Depending on the zoom level, this is either the full image or a sub-region.
- The colorbar
The colorbar shows the reference for the used colormap to map data levels to colors.
- The position information
This widget displays the coordinates and data values of the data under the mouse cursor.
Two-dimensional plots are presented in a silx Plot2D widget. The toolbar options will be explained in detail below. Moving the mouse over the canvas will update the labels for x/y position and data value at the bottom of the canvas. Note that the x and y axis positions for each pixel are defined at the pixel center and the given values must be treated carefully with respect to the pixel shape, especially for coarse pixels.
Tip
The scaling of the results can be achieved by modifying the colormap settings.